Find peaks (maxima) in a time series. This function is modified from
pracma::findpeaks.
findpeaks(
x,
nups = 1,
ndowns = nups,
zero = "0",
peakpat = NULL,
minpeakheight = -Inf,
minpeakdistance = 1,
h_min = 0,
h_max = 0,
npeaks = 0,
sortstr = FALSE,
include_gregexpr = FALSE,
IsPlot = F
)Arguments
- x
Numeric vector.
- nups
minimum number of increasing steps before a peak is reached
- ndowns
minimum number of decreasing steps after the peak
- zero
can be
+,-, or0; how to interprete succeeding steps of the same value: increasing, decreasing, or special- peakpat
define a peak as a regular pattern, such as the default pattern
[+]{1,}[-]{1,}; if a pattern is provided, the parametersnupsandndownsare not taken into account- minpeakheight
The minimum (absolute) height a peak has to have to be recognized as such
- minpeakdistance
The minimum distance (in indices) peaks have to have to be counted. If the distance of two maximum extreme value less than
minpeakdistance, only the real maximum value will be left.- h_min
his defined as the difference of peak value to the adjacent left and right trough value (h_leftandh_rightrespectively). The real peaks should followmin(h_left, h_right) >= h_min.- h_max
Similar as
h_min, the real peaks should followmax(h_left, h_right) >= h_min.- npeaks
the number of peaks to return. If
sortstr= true, the largest npeaks maximum values will be returned; Ifsortstr= false, just the first npeaks are returned in the order of index.- sortstr
Boolean, Should the peaks be returned sorted in decreasing oreder of their maximum value?
- include_gregexpr
Boolean (default
FALSE), whether to include the matchedgregexpr?- IsPlot
Boolean, whether to plot?
Note
In versions before v0.3.4, findpeaks(c(1, 2, 3, 4, 4, 3, 1)) failed to detect
peaks when a flat pattern exit in the middle.
From version v0.3.4, the peak pattern was changed from [+]{%d,}[-]{%d,} to
[+]{%d,}[0]{0,}[-]{%d,}. The latter can escape the flat part successfully.
Examples
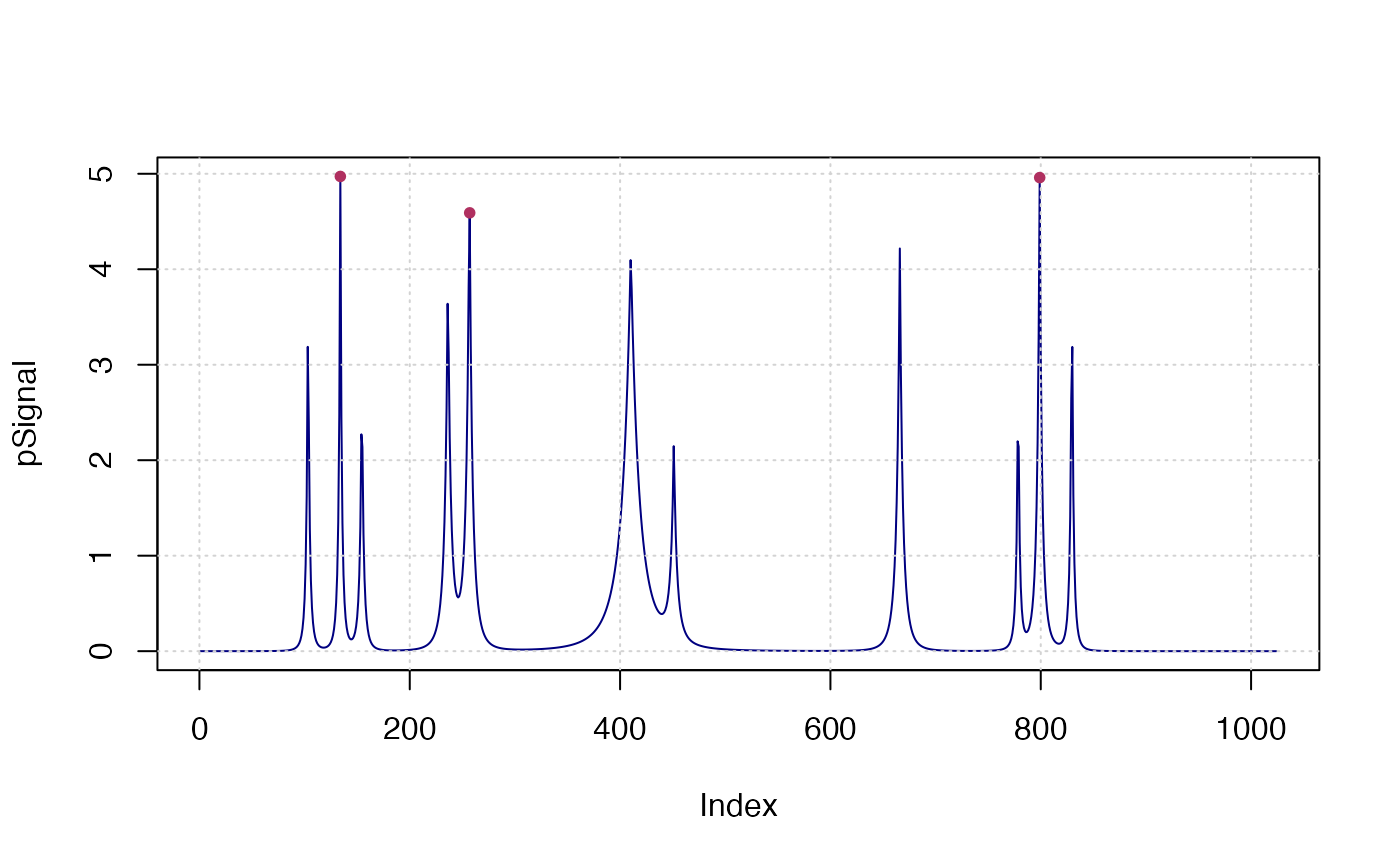
x <- seq(0, 1, len = 1024)
pos <- c(0.1, 0.13, 0.15, 0.23, 0.25, 0.40, 0.44, 0.65, 0.76, 0.78, 0.81)
hgt <- c(4, 5, 3, 4, 5, 4.2, 2.1, 4.3, 3.1, 5.1, 4.2)
wdt <- c(0.005, 0.005, 0.006, 0.01, 0.01, 0.03, 0.01, 0.01, 0.005, 0.008, 0.005)
pSignal <- numeric(length(x))
for (i in seq(along=pos)) {
pSignal <- pSignal + hgt[i]/(1 + abs((x - pos[i])/wdt[i]))^4
}
plot(pSignal, type="l", col="navy"); grid()
x <- findpeaks(pSignal, npeaks=3, h_min=4, sortstr=TRUE)
points(val~pos, x$X, pch=20, col="maroon")