I_optimx is rich of functionality, but with a low computing
performance. Some basic optimization functions are unified here, with some
input and output format.
opt_ncminfGeneral-Purpose Unconstrained Non-Linear Optimization, seeucminf::ucminf().opt_nlminbOptimization using PORT routines, seestats::nlminb().opt_nlmNon-Linear Minimization,stats::nlm().opt_optimGeneral-purpose Optimization, seestats::optim().
opt_ucminf(par0, objective, ...)
opt_nlm(par0, objective, ...)
opt_optim(par0, objective, method = "BFGS", ...)
opt_nlminb(par0, objective, ...)Arguments
- par0
Initial values for the parameters to be optimized over.
- objective
A function to be minimized (or maximized), with first argument the vector of parameters over which minimization is to take place. It should return a scalar result.
- ...
other parameters passed to
objective.- method
optimization method to be used in
p_optim. Seestats::optim().
Value
convcode: An integer code. 0 indicates successful convergence. Various methods may or may not return sufficient information to allow all the codes to be specified. An incomplete list of codes includes1: indicates that the iteration limitmaxithad been reached.20: indicates that the initial set of parameters is inadmissible, that is, that the function cannot be computed or returns an infinite, NULL, or NA value.21: indicates that an intermediate set of parameters is inadmissible.10: indicates degeneracy of the Nelder–Mead simplex.51: indicates a warning from the"L-BFGS-B"method; see componentmessagefor further details.52: indicates an error from the"L-BFGS-B"method; see componentmessagefor further details.9999: error
value: The value of fn corresponding to parpar: The best parameter foundnitns: the number of iterationsfevals: The number of calls toobjective.
See also
Examples
library(phenofit)
library(ggplot2)
library(magrittr)
library(purrr)
#>
#> Attaching package: ‘purrr’
#> The following object is masked from ‘package:magrittr’:
#>
#> set_names
library(data.table)
#>
#> Attaching package: ‘data.table’
#> The following object is masked from ‘package:purrr’:
#>
#> transpose
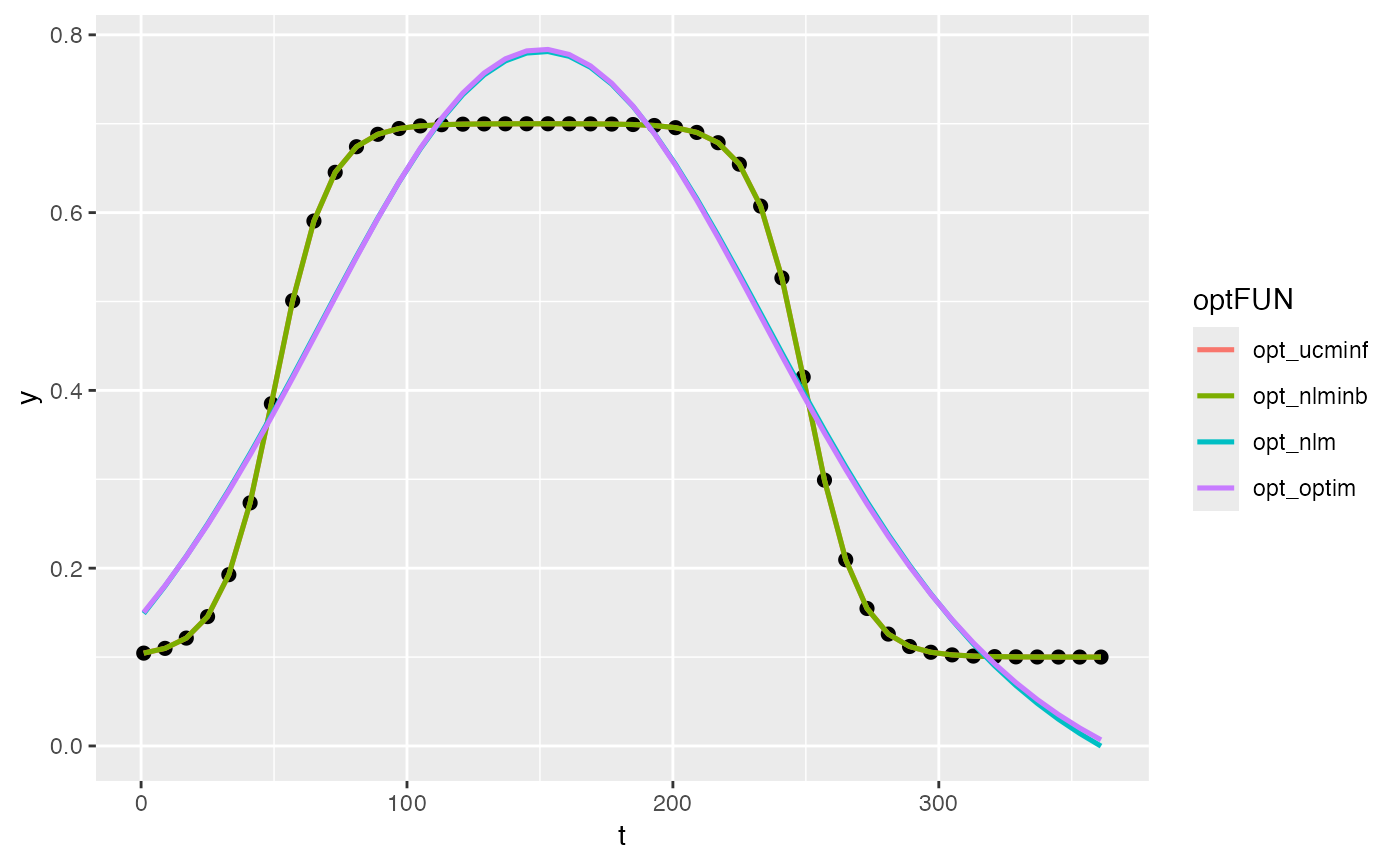
# simulate vegetation time-series
fFUN = doubleLog_Beck
par = c( mn = 0.1 , mx = 0.7 , sos = 50 , rsp = 0.1 , eos = 250, rau = 0.1)
par0 = c( mn = 0.15, mx = 0.65, sos = 100, rsp = 0.12, eos = 200, rau = 0.12)
t <- seq(1, 365, 8)
tout <- seq(1, 365, 1)
y <- fFUN(par, t)
optFUNs <- c("opt_ucminf", "opt_nlminb", "opt_nlm", "opt_optim") %>% set_names(., .)
opts <- lapply(optFUNs, function(optFUN){
optFUN <- get(optFUN)
opt <- optFUN(par0, f_goal, y = y, t = t, fun = fFUN)
opt$ysim <- fFUN(opt$par, t)
opt
})
# visualization
df <- map(opts, "ysim") %>% as.data.table() %>% cbind(t, y, .)
pdat <- data.table::melt(df, c("t", "y"), variable.name = "optFUN")
ggplot(pdat) +
geom_point(data = data.frame(t, y), aes(t, y), size = 2) +
geom_line(aes(t, value, color = optFUN), linewidth = 0.9)