Example
Load data
d = MOD13A1$dt %>% subset(site == "CA-NS6" & date >= "2010-01-01" & date <= "2016-12-31") %>%
.[, .(date, y = EVI/1e4, DayOfYear, QC = SummaryQA)]
d %<>% mutate(t = getRealDate(date, DayOfYear)) %>%
cbind(d[, as.list(qc_summary(QC, wmin = 0.2, wmid = 0.5, wmax = 0.8))]) %>%
.[, .(date, t, y, QC_flag, w)]
print(d)
#> date t y QC_flag w
#> 1: 2010-01-01 2010-01-08 0.1531 snow 0.2
#> 2: 2010-01-17 2010-01-25 0.1196 snow 0.2
#> 3: 2010-02-02 2010-02-13 0.1637 snow 0.2
#> 4: 2010-02-18 2010-02-25 0.1301 snow 0.2
#> 5: 2010-03-06 2010-03-21 0.1076 snow 0.2
#> ---
#> 157: 2016-10-15 2016-10-23 0.1272 snow 0.2
#> 158: 2016-10-31 2016-11-07 0.1773 snow 0.2
#> 159: 2016-11-16 2016-11-25 0.0711 cloud 0.2
#> 160: 2016-12-02 2016-12-12 0.1372 snow 0.2
#> 161: 2016-12-18 2017-01-02 0.1075 snow 0.2date: image date t : composite date
QC_flag and date are optional.
phenofit parameters
lambda <- 8
nptperyear <- 23
minExtendMonth <- 0.5
maxExtendMonth <- 1
minPercValid <- 0
wFUN <- wTSM # wBisquare
wmin <- 0.2
methods_fine <- c("AG", "Zhang", "Beck", "Elmore", "Gu")growing season division and fine-curve fitting
Simply treating calendar year as a complete growing season will
induce a considerable error for phenology extraction. A simple growing
season dividing method was proposed in phenofit.
The growing season dividing method rely on heavily in Whittaker smoother.
Procedures of initial weight, growing season dividing, curve fitting, and phenology extraction are conducted separately.
INPUT <- check_input(d$t, d$y, d$w,
QC_flag = d$QC_flag,
nptperyear = nptperyear,
maxgap = nptperyear / 4, wmin = 0.2
)
brks <- season_mov(INPUT,
list(FUN = "smooth_wWHIT", wFUN = wFUN,
maxExtendMonth = 3,
wmin = wmin, r_min = 0.1
))
# plot_season(INPUT, brks)
## 2.4 Curve fitting
fit <- curvefits(INPUT, brks,
list(
methods = methods_fine, # ,"klos",, 'Gu'
wFUN = wFUN,
iters = 2,
wmin = wmin,
# constrain = FALSE,
nextend = 2,
maxExtendMonth = maxExtendMonth, minExtendMonth = minExtendMonth,
minPercValid = minPercValid
))
## check the curve fitting parameters
l_param <- get_param(fit)
print(l_param$Beck)
#> # A tibble: 7 × 7
#> flag mn mx sos rsp eos rau
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2010_1 0.191 0.538 3808. 0.0826 3894. 0.0432
#> 2 2011_1 0.194 0.477 4180. 0.113 4274. 0.113
#> 3 2012_1 0.189 0.514 4542. 0.196 4643. 0.0864
#> 4 2013_1 0.190 0.493 4903. 0.190 5011. 0.0672
#> 5 2014_1 0.198 0.494 5276. 0.122 5375. 0.289
#> 6 2015_1 0.212 0.493 5639. 0.142 5726. 0.169
#> 7 2016_1 0.198 0.501 6002. 0.186 6095. 0.0731
dfit <- get_fitting(fit)
print(dfit)
#> flag t y QC_flag meth ziter1 ziter2
#> 1: 2010_1 2010-01-25 0.1196 snow AG 0.1888598 0.1903677
#> 2: 2010_1 2010-01-25 0.1196 snow Zhang 0.1881071 0.1897593
#> 3: 2010_1 2010-01-25 0.1196 snow Beck 0.1888971 0.1906256
#> 4: 2010_1 2010-01-25 0.1196 snow Elmore 0.1882156 0.1906010
#> 5: 2010_1 2010-01-25 0.1196 snow Gu 0.1876635 0.1878432
#> ---
#> 761: 2016_1 2016-12-12 0.1372 snow AG 0.1933491 0.1963926
#> 762: 2016_1 2016-12-12 0.1372 snow Zhang 0.1939335 0.1981793
#> 763: 2016_1 2016-12-12 0.1372 snow Beck 0.1940403 0.1985395
#> 764: 2016_1 2016-12-12 0.1372 snow Elmore 0.1941100 0.1986073
#> 765: 2016_1 2016-12-12 0.1372 snow Gu 0.1872110 0.1872110
## 2.5 Extract phenology
TRS <- c(0.1, 0.2, 0.5)
l_pheno <- get_pheno(fit, TRS = TRS, IsPlot = FALSE) # %>% map(~melt_list(., "meth"))
print(l_pheno$doy$Beck)
#> flag origin TRS1.sos TRS1.eos TRS2.sos TRS2.eos TRS5.sos TRS5.eos
#> 1: 2010_1 2010-01-01 126 300 137 280 154 248
#> 2: 2011_1 2011-01-01 143 278 151 270 163 257
#> 3: 2012_1 2012-01-01 148 288 153 277 160 261
#> 4: 2013_1 2013-01-01 142 298 147 284 154 262
#> 5: 2014_1 2014-01-01 143 271 151 267 163 262
#> 6: 2015_1 2015-01-01 144 262 150 256 161 248
#> 7: 2016_1 2016-01-01 147 285 151 272 159 253
#> DER.sos DER.pos DER.eos UD SD DD RD Greenup Maturity Senescence Dormancy
#> 1: 155 192 242 132 175 211 289 128 184 213 295
#> 2: 163 210 257 146 181 240 277 142 185 236 279
#> 3: 160 194 261 150 170 239 284 145 175 233 288
#> 4: 155 187 263 144 165 236 293 139 170 228 297
#> 5: 163 230 262 146 179 253 273 142 183 250 274
#> 6: 161 207 248 147 175 235 261 142 179 231 264
#> 7: 159 189 252 148 170 228 280 144 175 221 284
pheno <- l_pheno$doy %>% melt_list("meth")Visualization
# growing season dividing
plot_season(INPUT, brks, ylab = "EVI")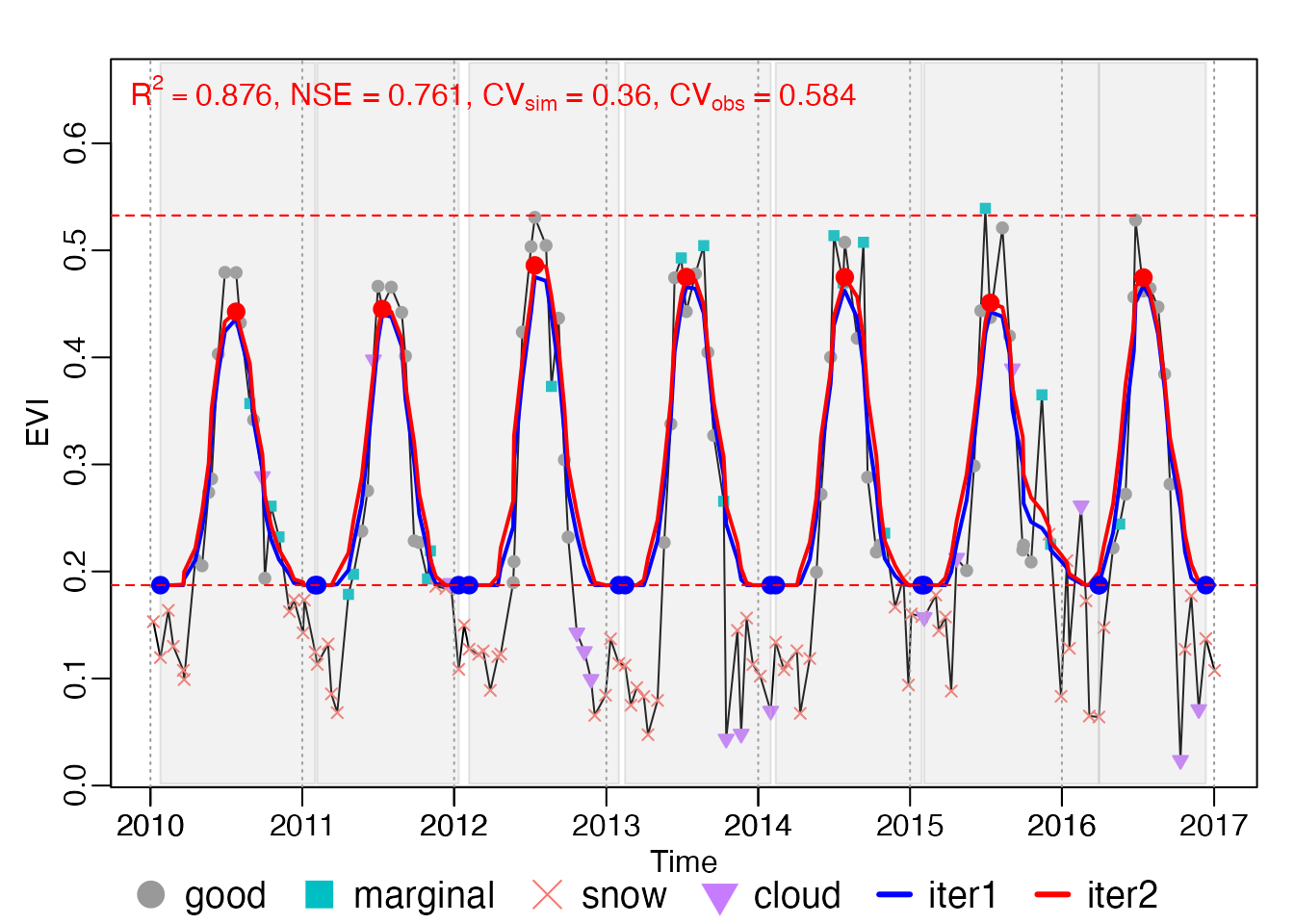
# Ipaper::write_fig({ }, "Figure4_seasons.pdf", 9, 4)
# fine curvefitting
g <- plot_curvefits(dfit, brks, title = NULL, cex = 1.5, ylab = "EVI", angle = 0)
grid::grid.newpage()
grid::grid.draw(g)
# Ipaper::write_fig(g, "Figure5_curvefitting.pdf", 8, 6, show = TRUE)
# extract phenology metrics, only the first 3 year showed at here
# write_fig({
l_pheno <- get_pheno(fit[1:7], method = "AG", TRS = TRS, IsPlot = TRUE, show.title = FALSE)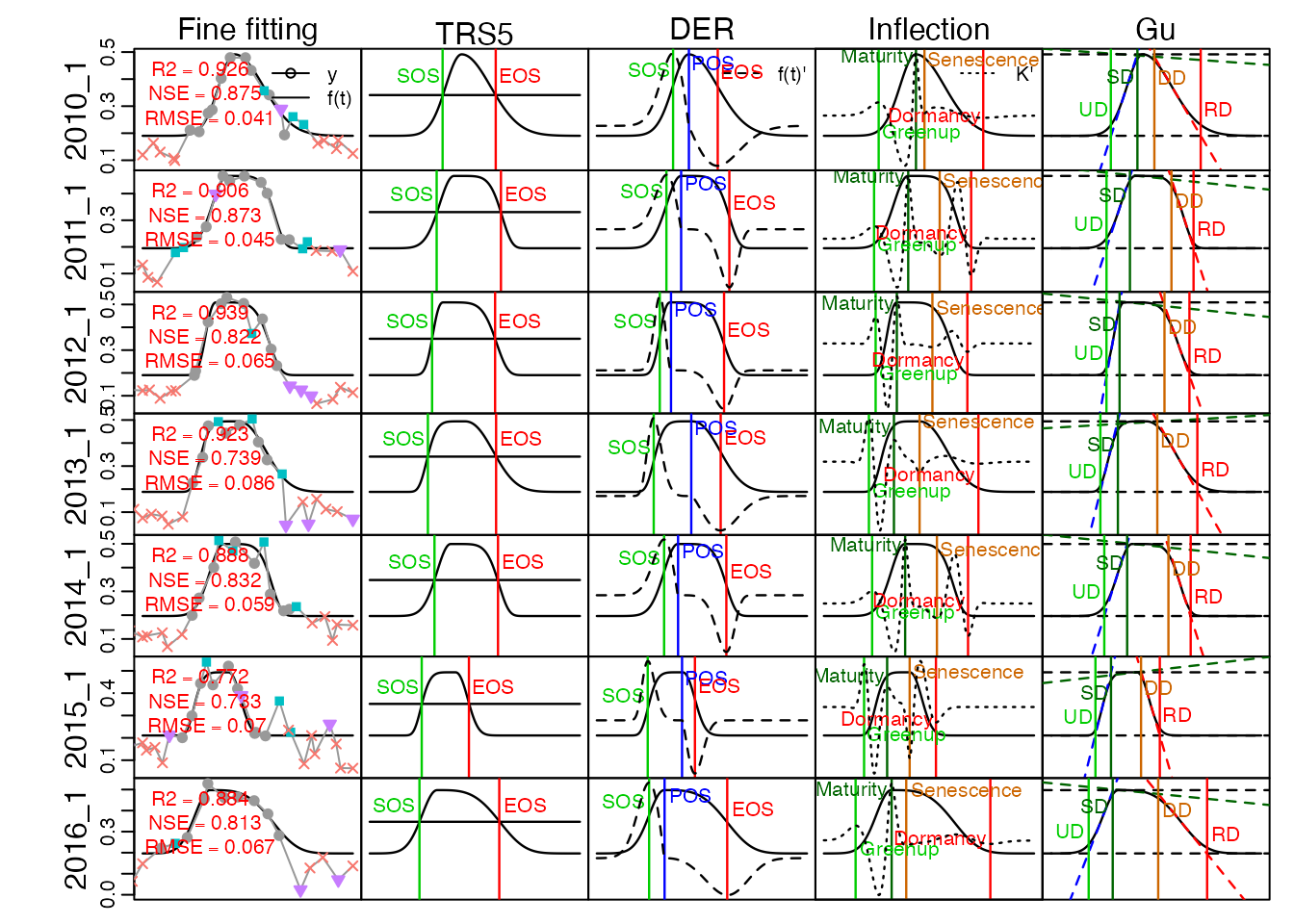
# }, "Figure6_phenology_metrics.pdf", 8, 6, show = TRUE)Comparison with TIMESAT and phenopix
# library(ggplot2)
# library(ggnewscale)
# # on the top of `Figure7_predata...`
# d_comp = fread("data-raw/dat_Figure7_comparison_with_others-CA-NS6.csv")
# d_comp = merge(d[, .(date, t)], d_comp[, .(date, TIMESAT, phenopix)]) %>%
# merge(dfit[meth == "Beck", .(t, phenofit = ziter2)], by = "t") %>%
# melt(c("date", "t"), variable.name = "meth")
# labels = c("good", "marginal", "snow", "cloud")
# theme_set(theme_grey(base_size = 16))
# cols_line = c(phenofit = "red", TIMESAT = "blue", phenopix = "black")
# p <- ggplot(dfit, aes(t, y)) +
# geom_point(aes(color = QC_flag, fill = QC_flag, shape = QC_flag), size = 3) +
# scale_shape_manual(values = qc_shapes[labels], guide = guide_legend(order = 1)) +
# scale_color_manual(values = qc_colors[labels], guide = guide_legend(order = 1)) +
# scale_fill_manual(values = qc_colors[labels], guide = guide_legend(order = 1)) +
# new_scale_color() +
# geom_line(data = d_comp, aes(t, value, color = meth)) +
# # geom_line(data = d_comp[meth == "phenofit"], aes(t, value),
# # size = 1, show.legend = FALSE, color = "red") +
# scale_color_manual(values = cols_line, guide = guide_legend(order = 2)) +
# labs(x = "Time", y = "EVI") +
# theme(
# axis.title.x = element_text(margin = margin(t = 0, unit='cm')),
# # plot.margin = margin(t = 0, unit='cm'),
# legend.text = element_text(size = 13),
# legend.position = "bottom",
# legend.title = element_blank(),
# legend.margin = margin(t = -0.3, unit='cm'))
# p
# # write_fig(p, "Figure7_comparison_with_others.pdf", 10, 4, show = TRUE)